
 |
understanding reverse rate calculation for chemical reactions
Hello all,
I'm trying to understand how the reverse rate is calculated. Following the manual of chemkin (https://www3.nd.edu/~powers/ame.60636/chemkin2000.pdf) the reverse rate of the ith reaction is calculated as follows: 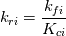 the equilibrium constant is calculated as follows: 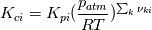 being 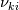 the sum of the stoichiometric coefficient of the ith reaction. Furthermore the sum of the stoichiometric coefficient of the ith reaction. Furthermore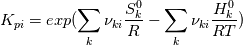 being 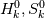 are the enthalpy and entropy of the species k. are the enthalpy and entropy of the species k.the source code for the calculation of k_ri can be found in $FOAM_SRC/thermophysicalModels/specie/reaction/Reactions/ReversibleReaction/ReversibleReaction.C: Code:
templateCode:
template<class Thermo, template<class> class Type>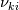 enter into the source code in openfoam. enter into the source code in openfoam. Best, Michael |
Thank you Adhiraj for pointing me in the right direction. Looking at the constructor of the Reaction class ($FOAM_SRC/thermophysicalModels/specie/reaction/Reactions/Reaction/Reaction.C)
we see the function setThermo Code:
template<class ReactionThermo>in this function we see that the stoichiometric coefficients and the molar weight of the species enter in the construction of the thermodynamic properties: Code:
template<class ReactionThermo> |
I had a look how the exponent in
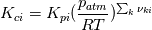 is calculated. is calculated. Key feature is the function setThermo in the file Reaction.C: Code:
To calculate the exponent we need to find out how this operation affect the Y_ and molWeight_ data of the of the thermodynamic object is affected by this operation. Since the thermodynamic object inherits from the equation of state object and this again inherits from the species object and the latter carries the data Y_ and molWeight_ we'll have a look how the above operators are defined in $FOAM_SRC/thermophysicalModels/specie/specie/specieI.H: Code:
inline specie operator*(const scalar s, const specie& st)Code:
molWeight_ = sumY/(Y_/molWeight_ + st.Y_/st.molWeight_);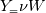 and 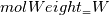 after the summation we obtain hence 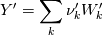 and 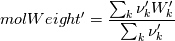 for the left hand side (the forward side of the reaction). An analogous formulation is obtained also for the right hand side. After the application of the == operator we obtain: 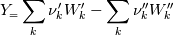 and 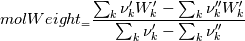 We see that the exponent nm is computed as follows: Code:
const scalar nm = this->Y()/this->W();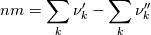 |
If someone is interest there is a description of how the reverse rate constant is calculated also here: https://openfoamwiki.net/index.php/ChemFoam
|
Hi mAlletto, then how should we explain the calculation of arg = -this->Y()*this->G(Pstd, T)/(RR*T) in OpenFoam instead of -this->G(Pstd, T)/(RR*T)?
Thanks. |
It is still not clear why Y=v*W
|
| All times are GMT -4. The time now is 15:38. |